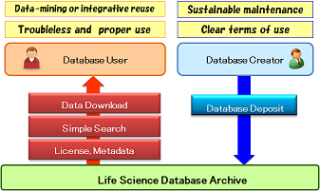
If a database is inadequate in terms of its description, unclear with respect to the terms of use, or is not downloadable, it may not be fully used, cited or rightly acknowledged by the (research) communities. This is even true for databases with high-quality datasets.
The Life Science Database Archive maintains and stores the datasets generated by life scientists in Japan in a long-term and stable state as national public goods. The Archive makes it easier for many people to search datasets by metadata (description of datasets) in a unified format, and to access and download the datasets with clear terms of use (see here for detailed descriptions).
In addition, the Archive provides datasets in forms friendly to different types of users in public and private institutions, and thereby supports further contribution of each research to life science.
| Database name⇅ | Maintenance site⇅ | Principal creator⇅ | Database classification⇅ | Organism⇅ | Summary⇅ | License⇅ |
|---|---|---|---|---|---|---|
| - | Akira Sonoda | marine flora and fauna, marine environment monitoring data | coral, fish, algae |
Images of organisms inhabiting the coral reef waters around the Ryukyu Islands provided by citizen divers and videos taken by GODAC's remotely operated mini-submarine “Niraikanai 150.” These were published on the Coral Reef Network Web System (http://www.godac.jp/coral/). |
CC BY-SA Detail |
|
| Biotechnology Center, National Institute for Technology and Evaluation | Natsuko Ichikawa | Metabolic and Signaling Pathways | Streptomyces |
Database of Streptomyces-derived Secondary Metabolites |
CC BY Detail |
|
| National Agriculture and Food Research Organization (NARO) | Akio Miyao | Plant databases | Rice |
Database of rice (Nipponbare) Tos17 insertion mutant lines |
CC BY-SA Detail |
|
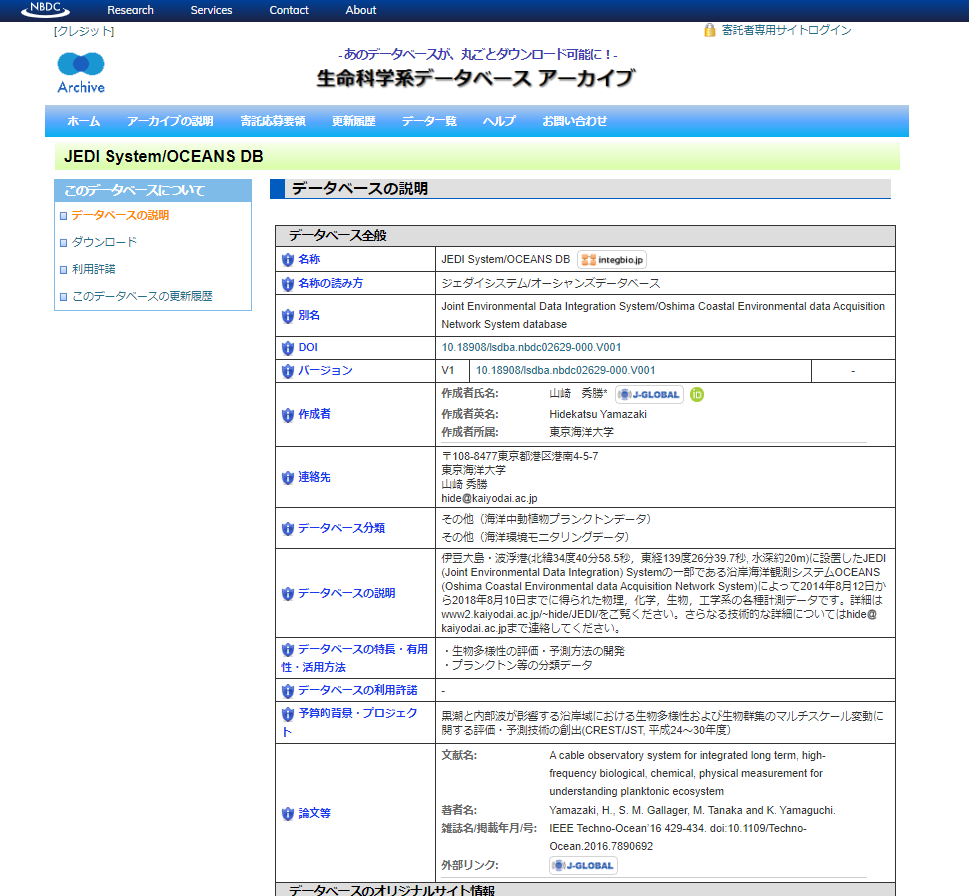
| - | Hidekatsu Yamasaki | Marine environmental monitoring data | Marine plankton |
The physical, chemical, biological and engineering data observed by OCEANS of JEDI System
|
- Detail |
|
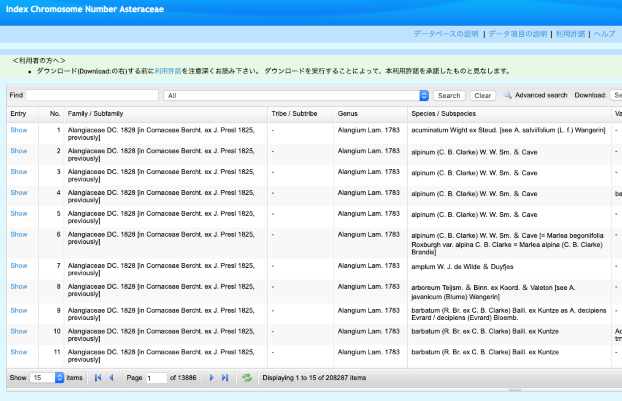
| - | Watanabe, Kuniaki | Literature | Asterales |
This database collects published data about Asterales needed for elucidating, classification, phylogeny, and evolution. |
CC BY-SA Detail |
|
| Database name⇅ | Maintenance site⇅ | Principal creator⇅ | Database classification⇅ | Organism⇅ | Summary⇅ | License⇅ |