Microarray isc.
|
| ‘ Functional Analysis of Genes |
|
@The functional analysis of genes is a way to find what roles the genes play in the living organism. It is important to understand what proteins the genes code, and where the genes are expressed (in tissues or organs) and when they are expressed. The methods for studying the expression of genes generally involve the expression at the transcription level. Following is a list of methods that can be used at the transcription level to analyze gene expression.
The macroarray method consists of three parts: fixing cloned genes on a support, such as nylon membrane, hybridizing the cloned genes and mRNA labeled radioactively with 33P (= labeled cDNA by reverse transcription), and taking autoradiography to examine the amount of gene expression.
|
| ‘ Principl of the Microarray Technique |
|
@The theory for microarrays is the same as for macroarrays. The differences are as follows: the gene fragments (probes) are fixed on a glass slide instead of a membrane support; the size and density of the arrays are smaller and higher. The analysis of genetic expression of the hybridized RNA (target) is not done with isotopes but with florescent dye markets (for details refer to the experimental ramification section). The number of probes and number of genes can be increased, and it is possible to analyze the gene expression of the comprised genes. Conversely, the probes used in the macroarrays and microarrays give no information on the expression of unfixed genes. Therefore, from the point of view of analyzing the expression of the genes in the genome, the number of probes required should be maximum. Currently there are two methods that are used for the microarrays.
|
| ‘ The GeneChipTM |
|
@This is totally different from a cDNA microarray spotting DNA clones on glass slide. For constructing the GeneChipTM, approximate 25-mer oligoDNAs, are directly synthesized on a glass slide using photolithography. Nucleotide sequence data are used to create a set of probes for each gene, which contain two types of probes: 25-mer oligoDNAs perfectly matching with 16 (~ 20) locations in each gene and the oligoDNAs of the same stretches except containing a single base substitution at the 13 th position of the 25 nucleotides. Once the chip is designed using data from published databases, it is possible to use the chip without maintaining and spotting DNA clones. The GeneChipTM is theoretically an ideal method for quantitative analysis of gene expression because the length and GC content of probes can be uniformed to make Tm value, the determinant for stringency of hybridization, even between probes. There are three drawbacks in the GeneChipTM method: it is impossible to construct the GeneChipTM in one's own laboratory; if there is no information about nucleotide sequence of a gene, it is impossible to make probes for the GeneChipTM; if one embark on further analysis about interensting genes based on the results of GeneChipTM experiment, it is necessary to isolate the DNA clones of genes because the probes are the short oligomers manufactured from information in nucleotide databases, not DNA clones. (Refer to the Affymetirix Company home page.)
|
| ‘ cDNA Microarray (= Microarray) |
| @cDNA microarray, the method to use cDNA clones fixed on glass slide as probes, is originally developed by the laboratory of Dr. Patrick O. Brown, Stanford University School of Medicine. There are various methods for spotting cDNA to glass slides, such as mechanical microspotting with capillary shaping pen(s) or noncapillary pen(s) (there are a variety of pen chapes) and ink jetting with miniature nozzle(s). The advantages of cDNA microarray system are that it is possible to prepare microarray at any time by spotting cDNA clones on glass slides and that all of the cDNA clones on microarray are already available to perform further analysis about interesting genes based on the results of microarray experiment. The disadvantages of these systems are that the equipments used for spotting and scannning and chemicals for labeling are expensive and that a large number of cDNA clones need to be prepared for spotting. |
|
| ‘ Potential Uses of Microarray |
|
@Microarray is a powerful tool for gene function analysis. For nearly all of the organisms whose genome are now being or have been sequenced, the cDNA sequencing analyses have also simultaneously been in progress or done. The functions for most of the genes are still unknown though some of genes show the similarities to the genes/cDNAs with known functions by the results of homology search in the databases based on cDNA sequences. The microarray technique allows one to study how the transcription levels of all genes on microarray change between different biological conditions, with a small number of repeated experiments. (Use of microarray for gene expression profiling) The technique also provide ways to look for the DNA clones that show the transcriptional changes under different conditions regarding a phenomenon that researchers are interested in (for example, response to environmental/biological stress, gene expression specificity in tissue/developmental stage, etc.), from all gene/cDNA clones on microarray. (Use of microarray as method to identify interesting/useful genes) In short, the microarray technique is a method to obtain very large amounts of gene expression data with a minimum number of experiments. @Generally speaking, the experimental techniques in molecular biology have been used to the biological phenomena whose genetic analyses are easy to perform and/or the cDNAs/genes involved in are easy to isolate. If the microarray technique apply to the interesting biological phenomena that are difficult to elucidate, such as heterosis (= hibrid vigor) and the totipotency of cultured plant cell, the breakthrough in molecular biology on these phenomena will occur
|
| ‘ Microarray as primary differential screening method |
| As mentioned above, the microarray technique is a method to obtain a large quantity of gene expression data with an experiment and to identify interesting/useful genes. However it should be stressed that "the data from microarray experiments do not provide all the answers. The microarray is a simple and quick method for determining which of the large number of genes on microarray should be selected for further analysis." By microarray experiment data, it is possible to detect the significant differences of transcription levels, which is generally more than twofold (or threefold), of many genes between different samples. But it is difficult to conclude that all of the differences are authentic, based on only microarray data. To resolve this difficulty, there are the follwoing issues: how to standardize the data, how to normalize the data obtained from differrent glass slides, how to set up positive/negative controls and use their values for data analysis, how to find the housekeeping genes showing constant expression levels. Although standardizing the data is possible in a few cases, that would be extremely difficult in the cases which show low expression of most of the genes in an organism, such as the states showing cell death in most of the cells. Because the perfect solving those issues pointed out above is practically impossible, the microarray should be regarded as primary screening method. And it would be advantageous to use the Northern hybridization method or the RT-PCR method for re-examining the levels of transcription for each gene selected by microarray data. |
|
|
|
|
@The microarray technique has been disseminated very rapidly. Consequently, there is little past history, and the best way to use the technique is still being debated. Following are several of these dispute areas.
iPjShould the two-dye or one-dye method be used? |
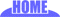


 Technical Background
Technical Background