Description
The MOUSE BASEMENT MEMBRANE BODYMAP is the first body-wide database of ECM proteins and
provides a bird's-eye view of the cell- and tissue-specific customization of the extracellular
microenvironment.
The MOUSE BASEMENT MEMBRANE BODYMAP contains a bodymap of 42 ECM proteins including 36 basement membrane proteins,
of which 31 are known proteins such as laminins, type IV collagens, nidogens and perlecan,
and 5 are newly identified proteins within this project, in whole mouse embryos on
standardized experimental platforms.
The MOUSE BASEMENT MEMBRANE BODYMAP consists of hundreds of high resolution virtual slides
representing the localization of basement membrane proteins in whole mouse embryos analyzed
by immunohistochemical technique. The virtual slides have been created by converting
gigapixel digital images produced from the immunohistochemistry data to ZOOMATM images
that allow you to enlarge, reduce and move the images as you want. The characteristic
features of the virtual slides in the MOUSE BASEMENT MEMBRANE BODYMAP enable you to
observe the protein localizations in whole mouse embryo slices more effectively than
using a conventional microscope.
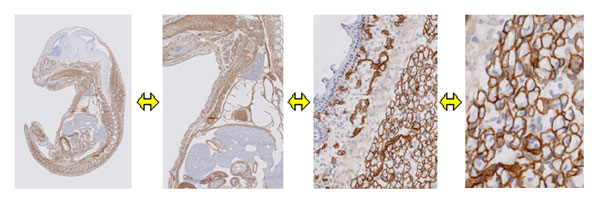
You can freely zoom and move the digital images, which were created from
immunohistochemically stained whole mouse embryo slices.
Another characteristic is that the dataset includes hematoxylin/eosin stained data.
Immunohistochemically stained sections often do not contain enough information to
identify cell/tissue types. In contrast, hematoxylin/eosin staining visualizes almost
all nuclei and proteins with different intensity that is dependent on cell/tissue types.
To facilitate the identification of cell/tissue types and the annotation of protein localizations,
we performed hematoxylin/eosin staining of all sections adjacent to the immunostained sections.
Each hematoxylin/eosin stained section was converted to a virtual slide, and zoom and positional
movement functions were synchronized with the virtual slide created from the corresponding
immunostained section. Thus, you can easily compare the immunostained data with the
corresponding hematoxylin/eosin stained data at any time during the observation.
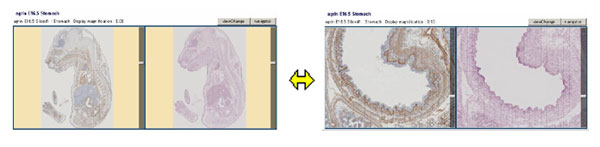
Each immunostained slice (left panel) has a hematoxylin/eosin stained adjacent
slice (right panel). ZOOM and positional movement functions of the digital images
are synchronized in the paired virtual slides.
|